Mapping the seasonal changes to the open water extent of the Okavango delta¶
Getting started¶
To run this analysis, run all the cells in the notebook, starting with the « Load packages » cell.
Load packages¶
Import Python packages that are used for the analysis.
[1]:
%matplotlib inline
# Force GeoPandas to use Shapely instead of PyGEOS
# In a future release, GeoPandas will switch to using Shapely by default.
import os
os.environ['USE_PYGEOS'] = '0'
import datacube
import matplotlib.pyplot as plt
import numpy as np
import sys
import xarray as xr
import pandas as pd
import geopandas as gpd
from IPython.display import Image
from matplotlib.colors import ListedColormap
from matplotlib.patches import Patch
from odc.ui import image_aspect
from datacube.utils import geometry
from datacube.utils.cog import write_cog
from deafrica_tools.datahandling import load_ard
from deafrica_tools.bandindices import calculate_indices
from deafrica_tools.plotting import map_shapefile
from deafrica_tools.dask import create_local_dask_cluster
from deafrica_tools.spatial import xr_rasterize
from datacube.utils.aws import configure_s3_access
configure_s3_access(aws_unsigned=True, cloud_defaults=True)
Set up a Dask cluster¶
Dask can be used to better manage memory use and conduct the analysis in parallel. For an introduction to using Dask with Digital Earth Africa, see the Dask notebook.
Note: We recommend opening the Dask processing window to view the different computations that are being executed; to do this, see the Dask dashboard in DE Africa section of the Dask notebook.
To activate Dask, set up the local computing cluster using the cell below.
[ ]:
create_local_dask_cluster()
Analysis parameters¶
The following cell sets the parameters, which define the area of interest and the length of time to conduct the analysis over.
The parameters are: * vector_file: The path to the shapefile or geojson that will define the analysis area of the study * products: The products to load from the datacube, e.g. 's2_l2a`, or`”ls8_sr”*time_range: The date range to analyse (e.g.(“2017”,
“2019”). *measurements: The spectral bands to load from the satellite product.MNDWIrequires the’green”and’swir_1”bands *resolution: The pixel resolution of the satellite data.(-30,30)for Landsat or(-10,10)for Sentinel-2 *dask_chunks`: Chunk sizes to use for dask, the default values below are optimized for the full Okavango delta
If running the notebook for the first time, keep the default settings below. This will demonstrate how the analysis works and provide meaningful results. The default area is the Ruko Conservancy.
[3]:
vector_file = 'data/okavango_delta_outline.geojson'
products = ['ls7_sr','ls8_sr']
wetness_index = 'MNDWI'
time_range = ('2013-12', '2021-05')
measurements = ['green','swir_1']
resolution = (-30,30)
dask_chunks = {'time':1,'x':1500,'y':1500}
View the Area of Interest on an interative map¶
The next cell will first open the vector file and then display the selected area on an interactive map. Zoom in and out to get a better understanding of the area of interest.
[4]:
#read shapefile
gdf = gpd.read_file(vector_file)
map_shapefile(gdf, attribute='GRID_CODE')
Connect to the datacube¶
Activate the datacube database, which provides functionality for loading and displaying stored Earth observation data.
[5]:
dc = datacube.Datacube(app='water_extent')
Load cloud-masked Satellite data¶
The first step is to load satellite data for the specified area of interest and time range.
[6]:
bbox=list(gdf.total_bounds)
lon_range = (bbox[0], bbox[2])
lat_range = (bbox[1], bbox[3])
#create the dc query
water_query = {
'x': lon_range,
'y': lat_range,
'time': time_range,
'measurements': measurements,
'resolution': resolution,
'output_crs': 'EPSG:6933',
'dask_chunks': dask_chunks,
'group_by':'solar_day'
}
Now load the satellite data
[7]:
ds = load_ard(dc=dc,
products=products,
**water_query
)
print(ds)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls7_sr
ls8_sr
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Returning 934 time steps as a dask array
<xarray.Dataset>
Dimensions: (time: 934, y: 7774, x: 6955)
Coordinates:
* time (time) datetime64[ns] 2013-12-01T08:33:19.653638 ... 2021-05...
* y (y) float64 -2.279e+06 -2.279e+06 ... -2.512e+06 -2.513e+06
* x (x) float64 2.097e+06 2.097e+06 ... 2.305e+06 2.305e+06
spatial_ref int32 6933
Data variables:
green (time, y, x) float32 dask.array<chunksize=(1, 1500, 1500), meta=np.ndarray>
swir_1 (time, y, x) float32 dask.array<chunksize=(1, 1500, 1500), meta=np.ndarray>
Attributes:
crs: EPSG:6933
grid_mapping: spatial_ref
Mask the satellite data with vector file¶
[8]:
#create mask
mask = xr_rasterize(gdf,ds)
#mask data
ds = ds.where(mask)
Calculate the wetness index¶
[9]:
# Calculate the chosen vegetation proxy index and add it to the loaded data set
ds = calculate_indices(ds=ds, index=[wetness_index], drop=True, satellite_mission='ls')
#convert to float 32 to conserve memory
ds = ds.astype(np.float32)
Dropping bands ['green', 'swir_1']
Resample time series¶
Due to many factors (e.g. cloud obscuring the region, missed cloud cover in the SCL layer) the data will be gappy and noisy. Here, we will resample the data to ensure we working with a consistent time-series.
To do this we resample the data to seasonal time-steps using medians
These calculations will take nearly an hour to complete as we will run .compute(), triggering all the tasks we scheduled above and bringing the arrays into memory.
[10]:
%%time
sample_frequency="QS-DEC" # quarterly starting in DEC, i.e. seasonal
#resample MNDWI using medians
print('calculating '+wetness_index+' seasonal medians...')
ds = ds.resample(time=sample_frequency).median("time").compute()
# Update the time coordinates of the resampled dataset.
ds = ds.assign_coords(time=((pd.to_datetime(ds.time) + pd.tseries.offsets.DateOffset(months=1)) - pd.tseries.offsets.DateOffset(days=1)))
calculating MNDWI seasonal medians...
/usr/local/lib/python3.10/dist-packages/rasterio/warp.py:344: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
_reproject(
CPU times: user 14min 7s, sys: 44.6 s, total: 14min 52s
Wall time: 1h 41min 14s
Facet plot the water extent for every season¶
[11]:
ds[wetness_index].plot.imshow(col='time', col_wrap=4, cmap='Blues', vmax=0.3, vmin=-0.5);
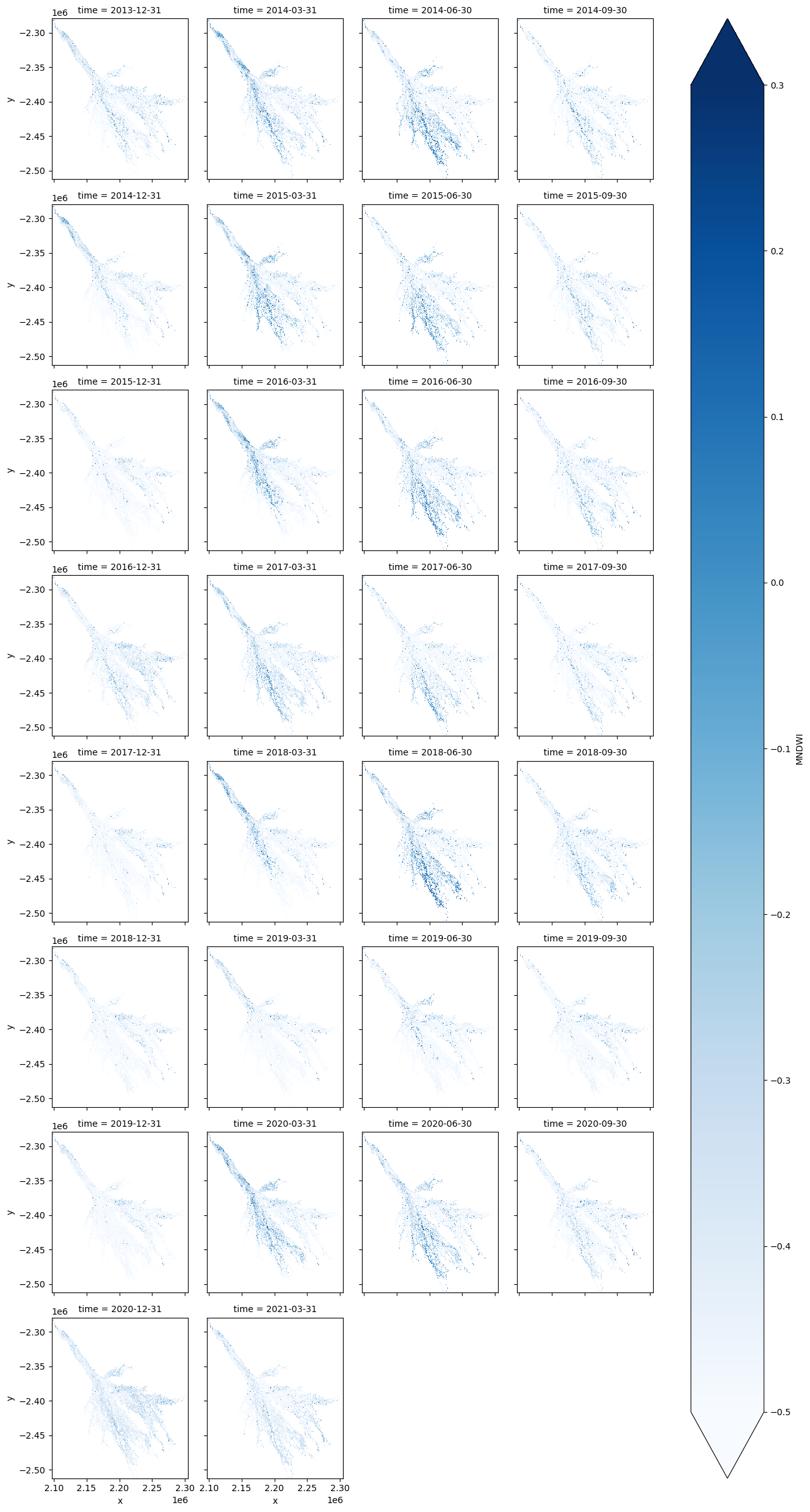
Calculate the water extent per time-step¶
The number of pixels can be used for the area of the waterbody if the pixel area is known. Run the following cell to generate the necessary constants for performing this conversion.
[12]:
pixel_length = water_query["resolution"][1] # in metres
m_per_km = 1000 # conversion from metres to kilometres
area_per_pixel = pixel_length**2 / m_per_km**2
Calculating the extent of open water¶
Calculates the area of pixels classified as water (if MNDWI is > 0, the water)
[13]:
water = ds[wetness_index].where(ds[wetness_index] > 0, np.nan)
area_ds = water.where(np.isnan(water), 1)
ds_valid_water_area = area_ds.sum(dim=['x', 'y']) * area_per_pixel
Export time-series as csv¶
[14]:
ds_valid_water_area.to_dataframe().drop('spatial_ref',axis=1).rename({'MNDWI':'Area of waterbodies (km2)'},axis=1).to_csv(f'results/water_extent_{time_range[0]}_to_{time_range[1]}.csv')
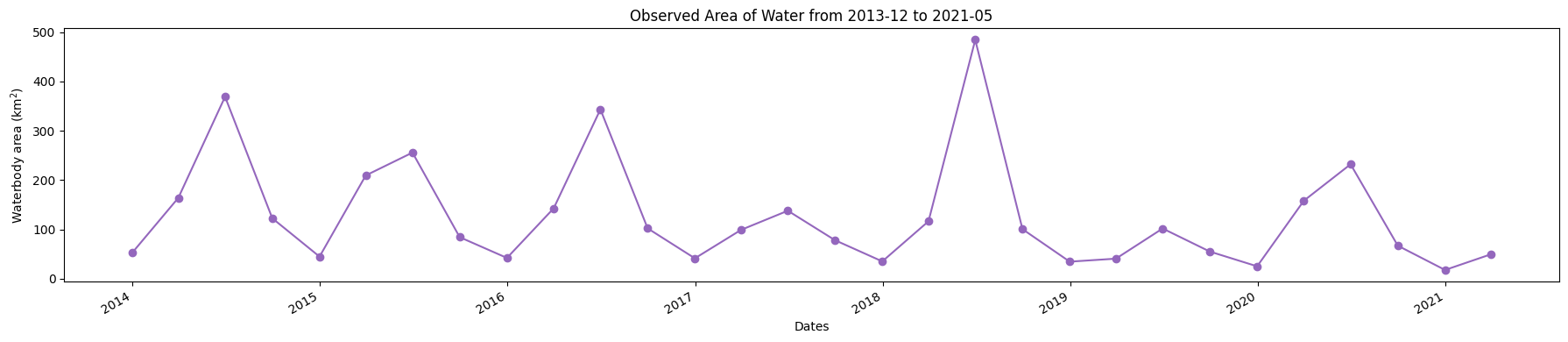
Plot a time series of open water area¶
[15]:
plt.figure(figsize=(18, 4))
ds_valid_water_area.plot(marker='o', color='#9467bd')
plt.title(f'Observed Area of Water from {time_range[0]} to {time_range[1]}')
plt.xlabel('Dates')
plt.ylabel('Waterbody area (km$^2$)')
plt.tight_layout()
plt.savefig(f'results/water_extent_{time_range[0]}_to_{time_range[1]}.png')
Compare water extent between two periods¶
baseline_time: The baseline year for the analysisanalysis_time: The year to compare to the baseline year
[16]:
baseline_time = '2018-06-30'
analysis_time = '2019-06-30'
[17]:
baseline_ds, analysis_ds = ds_valid_water_area.sel(time=baseline_time, method ='nearest'),ds_valid_water_area.sel(time=analysis_time, method ='nearest')
time_xr = xr.DataArray([baseline_ds.time.values, analysis_ds.time.values], dims=["time"])
print(time_xr.values)
['2018-06-30T00:00:00.000000000' '2019-06-30T00:00:00.000000000']
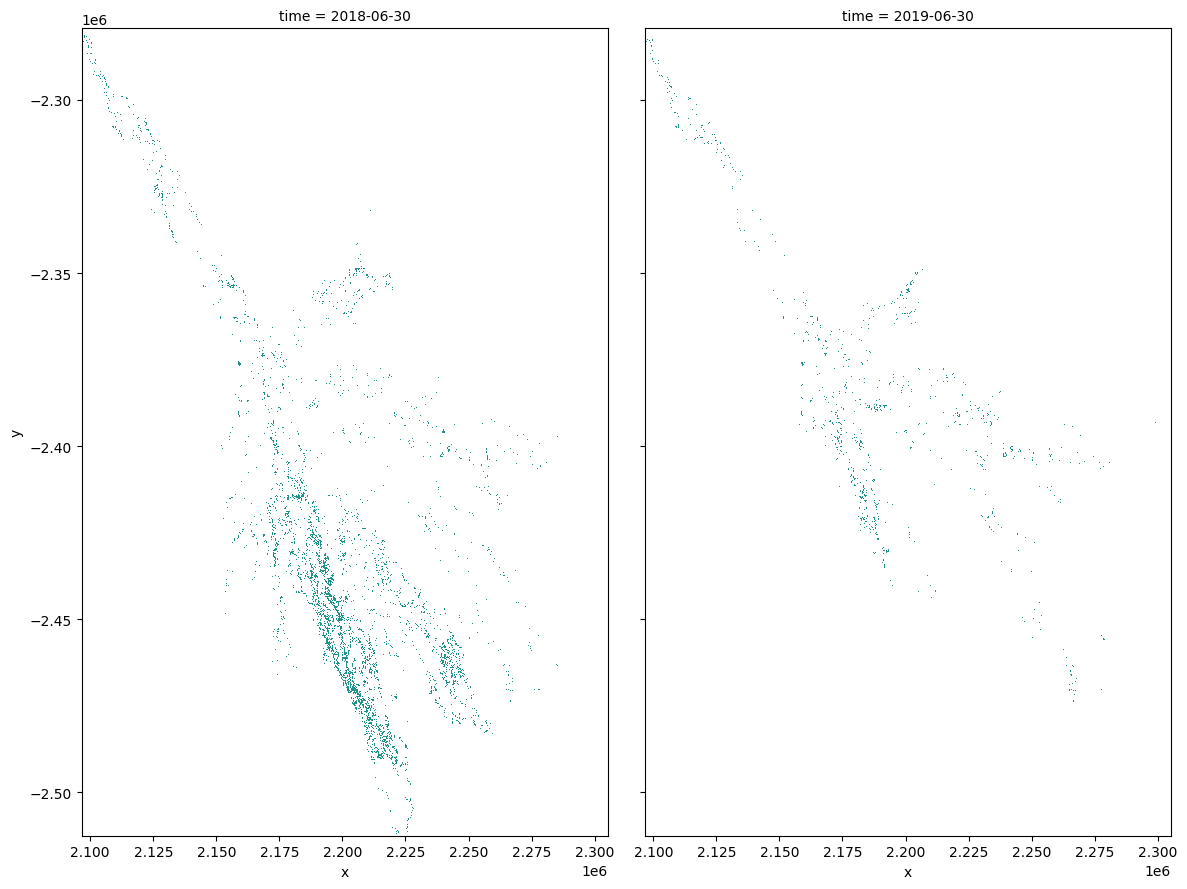
Plotting¶
Plot water extent of the MNDWI product for the two chosen periods.
[18]:
area_ds.sel(time=time_xr).plot(col="time", col_wrap=2, robust=True, figsize=(12, 9), cmap='viridis', add_colorbar=False);
Save the water extents as geotiffs¶
Both the “analysis time” and the “baseline time” water extents will be saved as cloud-opimtised geotiffs in the results/ folder
[19]:
write_cog(area_ds.sel(time=baseline_time),
fname='results/water_extent_'+baseline_time+'.tif',
overwrite=True)
write_cog(area_ds.sel(time=analysis_time),
fname='results/water_extent_'+analysis_time+'.tif',
overwrite=True)
[19]:
PosixPath('results/water_extent_2019-06-30.tif')
Calculate the change between the two nominated periods¶
The cells below calculate the amount of water gain, loss and stable for the two periods
[20]:
# The two period Extract the two periods(Baseline and analysis) dataset from
ds_selected = area_ds.where(area_ds == 1, 0).sel(time=time_xr)
analyse_total_value = ds_selected[1]
change = analyse_total_value - ds_selected[0]
water_appeared = change.where(change == 1)
permanent_water = change.where((change == 0) & (analyse_total_value == 1))
permanent_land = change.where((change == 0) & (analyse_total_value == 0))
water_disappeared = change.where(change == -1)
The cell below calculate the area of water extent for water_loss, water_gain, permanent water and land
[21]:
total_area = analyse_total_value.count().values * area_per_pixel
water_apperaed_area = water_appeared.count().values * area_per_pixel
permanent_water_area = permanent_water.count().values * area_per_pixel
water_disappeared_area = water_disappeared.count().values * area_per_pixel
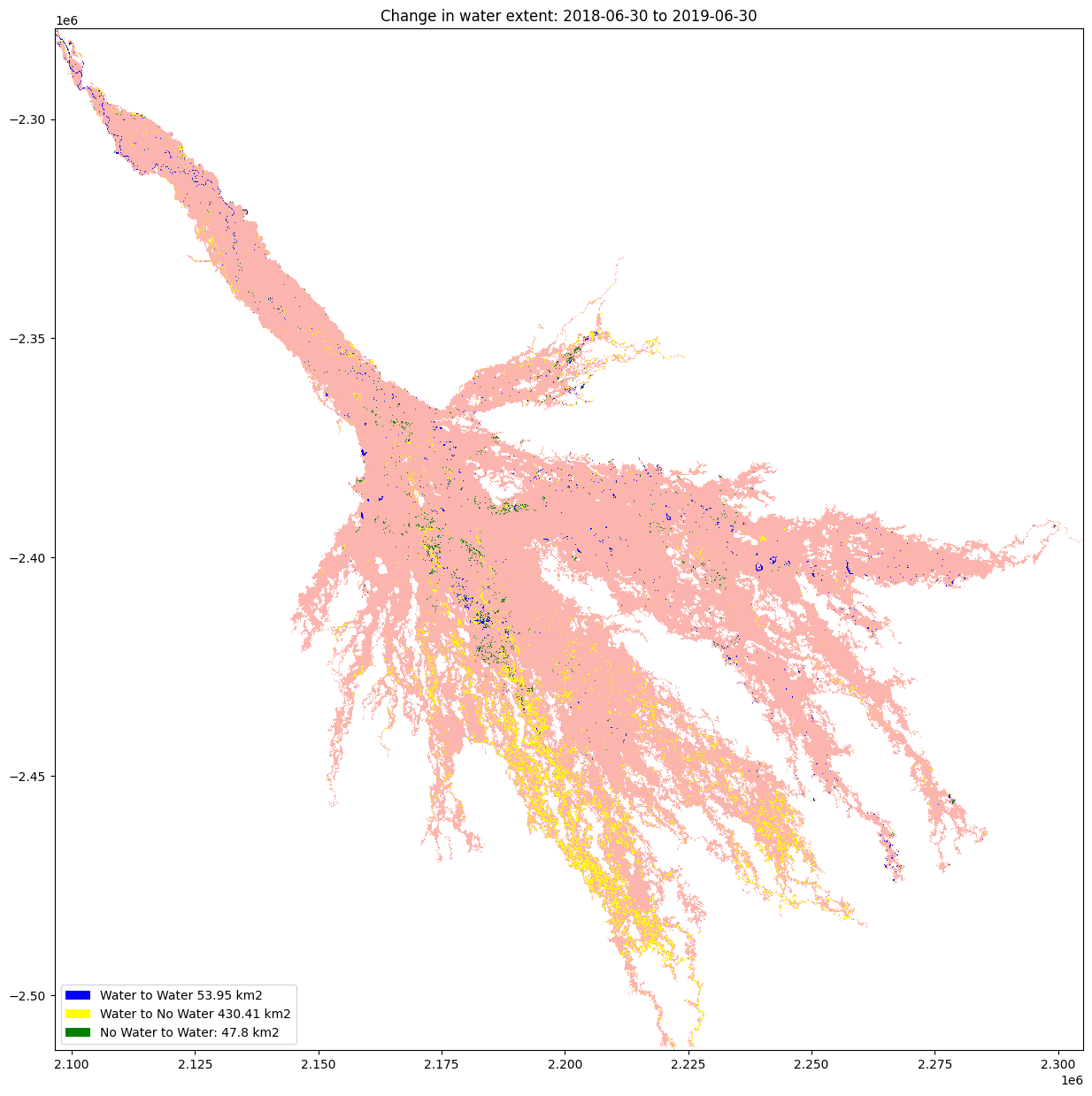
Plot the changes¶
The water variables are plotted to visualised the result
[22]:
water_appeared_color = "Green"
water_disappeared_color = "Yellow"
stable_color = "Blue"
land_color = "Brown"
fig, ax = plt.subplots(1, 1, figsize=(15, 15))
ds_selected[1].where(mask).plot.imshow(cmap="Pastel1",
add_colorbar=False,
add_labels=False,
ax=ax)
water_appeared.plot.imshow(
cmap=ListedColormap([water_appeared_color]),
add_colorbar=False,
add_labels=False,
ax=ax,
)
water_disappeared.plot.imshow(
cmap=ListedColormap([water_disappeared_color]),
add_colorbar=False,
add_labels=False,
ax=ax,
)
permanent_water.plot.imshow(cmap=ListedColormap([stable_color]),
add_colorbar=False,
add_labels=False,
ax=ax)
plt.legend(
[
Patch(facecolor=stable_color),
Patch(facecolor=water_disappeared_color),
Patch(facecolor=water_appeared_color),
Patch(facecolor=land_color),
],
[
f"Water to Water {round(permanent_water_area, 2)} km2",
f"Water to No Water {round(water_disappeared_area, 2)} km2",
f"No Water to Water: {round(water_apperaed_area, 2)} km2",
],
loc="lower left",
)
plt.title("Change in water extent: " + baseline_time + " to " + analysis_time);
Additional information¶
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Africa data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Slack channel or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
Github.
Compatible datacube version:
[23]:
print(datacube.__version__)
1.8.15
Last Tested:
[24]:
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
[24]:
'2023-08-11'