Using load_ard to load and cloud mask multiple satellite sensors¶
Keywords data used; landsat 5, data used; landsat 7, data used; landsat 8, data used; landsat 9, data used; sentinel-2, cloud masking, cloud filtering, pixel quality, dask
Description¶
This notebook demonstrates how to use the load_ard function to import a time series of cloud-free observations over Africa from multiple Landsat satellites (i.e. Landsat 5, 7, 8, and 9) and Sentinel-2 satellites (i.e. Sentinel-2A and Sentinel-2B). The function automatically applies cloud masking to the input data and returns all available data from multiple sensors as a single combined xarray.Dataset.
Optionally, the function can be used to return only observations that contain a minimum proportion of good quality non-cloudy or shadowed pixels. This can be used to extract visually appealing time series of observations that are not affected by cloud.
The load_ard function currently supports the following products:
USGS Collection 2:
ls5_sr, ls7_sr, ls8_sr, ls9_srSentinel 2:
s2_l2aSentinel 1:
s1_rtc
This notebook demonstrates how to use load_ard to 1. Load and combine Landsat 5, 7, 8, and 9 data into a single xarray.Dataset 2. Optionally apply a cloud mask to the resulting data 3. Filter resulting data to keep only cloud-free observations 4. Applying morphological processing on the cloud mask 5. Filter data before loading using a custom function 6. Load Sentinel-2 data 7. Lazily load data using Dask
Getting started¶
To run this analysis, run all the cells in the notebook, starting with the « Load packages » cell.
Load packages¶
[1]:
%matplotlib inline
import datacube
import matplotlib.pyplot as plt
from deafrica_tools.datahandling import load_ard
from deafrica_tools.plotting import rgb
Connect to the datacube¶
[2]:
# Connect to datacube
dc = datacube.Datacube(app='Using_load_ard')
Loading multiple Landsat sensors¶
The load_ard function can be used to load a single, combined timeseries of cloud-masked data from multiple Landsat satellites. At its simplest, you can use the function similarly to dc.load by passing a set of spatiotemporal query parameters (e.g. x, y, time, measurements, output_crs, resolution, group_by etc) directly into the function (see the dc.load documentation for all possible
options). The key difference from dc.load is that the function also requires an existing Datacube object, which is passed using the dc parameter. This gives us the flexibilty to load data from development or experimental datacubes.
In the examples below, we load three bands of data (red, green, blue) from the three USGS Collection 2 products (Landsat 5, 7, 8, and 9) by specifying:
products=['ls5_sr', 'ls7_sr', 'ls8_sr', ls9_sr']
Note: There are some limitations to
load_arddue to its design. An important part of its intended functionality is to provide a simple means of concatenating Landsat 5, 7, 8, and 9 together to form a single xarray object. However, this means thatload_ardcan only apply pixel quality masking to pq-categories that are common across Landsat sensors. For example, Landsat-8 has a dedicated bit flag for cirrus bands, but Landsat 5 and 7 do not. This means thatload_ardcannot accept'cirrus': 'high_confidence'as a pq-category. The same issue also applies for masking of aerosols, Landat 5 and 7 have different means of recording high aerosol than Landsat 8, and thusload_arddoes not support masking of aerosols.
The function outputs the number of observations for each product, and the total number loaded if the verbose= parameter is set to True (which it is by default)
Explicit syntax¶
The following example demonstrates how key parameters can be passed directly to load_ard.
[3]:
ds = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr',
'ls9_sr'],
x=(-16.45, -16.6),
y=(13.675, 13.8),
time=('2021-09', '2021-12'),
measurements = ['red','green','blue'],
output_crs='epsg:6933',
resolution=(-30, 30),
group_by='solar_day')
print(ds)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
ls9_sr
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Loading 17 time steps
/usr/local/lib/python3.10/dist-packages/rasterio/warp.py:344: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
_reproject(
<xarray.Dataset>
Dimensions: (time: 17, y: 517, x: 484)
Coordinates:
* time (time) datetime64[ns] 2021-09-05T11:27:43.160264 ... 2021-12...
* y (y) float64 1.744e+06 1.744e+06 ... 1.729e+06 1.728e+06
* x (x) float64 -1.602e+06 -1.602e+06 ... -1.587e+06 -1.587e+06
spatial_ref int32 6933
Data variables:
red (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
green (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
blue (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
Attributes:
crs: epsg:6933
grid_mapping: spatial_ref
Query syntax¶
The following example demonstrates how key parameters can be stored in a query dictionary, to be passed as a single parameter to load_ard. The query can then be reused in other load_ard calls.
[4]:
# Create a reusable query
query = {
'x': (-16.45, -16.6),
'y': (13.675, 13.8),
'time': ('2021-09', '2021-12'),
'measurements': ['red', 'green', 'blue'],
'group_by': 'solar_day',
'output_crs' : 'epsg:6933',
}
[5]:
# Load available data from all three Landsat satellites
ds = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr',
'ls9_sr'],
resolution=(-30, 30),
**query)
# Print output data
print(ds)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
ls9_sr
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Loading 17 time steps
<xarray.Dataset>
Dimensions: (time: 17, y: 517, x: 484)
Coordinates:
* time (time) datetime64[ns] 2021-09-05T11:27:43.160264 ... 2021-12...
* y (y) float64 1.744e+06 1.744e+06 ... 1.729e+06 1.728e+06
* x (x) float64 -1.602e+06 -1.602e+06 ... -1.587e+06 -1.587e+06
spatial_ref int32 6933
Data variables:
red (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
green (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
blue (time, y, x) float32 nan nan nan nan nan ... nan nan nan nan
Attributes:
crs: epsg:6933
grid_mapping: spatial_ref
Working with cloud masking¶
By plotting a time slice from the data we loaded above, you can see an area of white pixels where clouds have been masked out and set to NaN:
[6]:
# Plot single observation
rgb(ds, col='time', col_wrap=6)
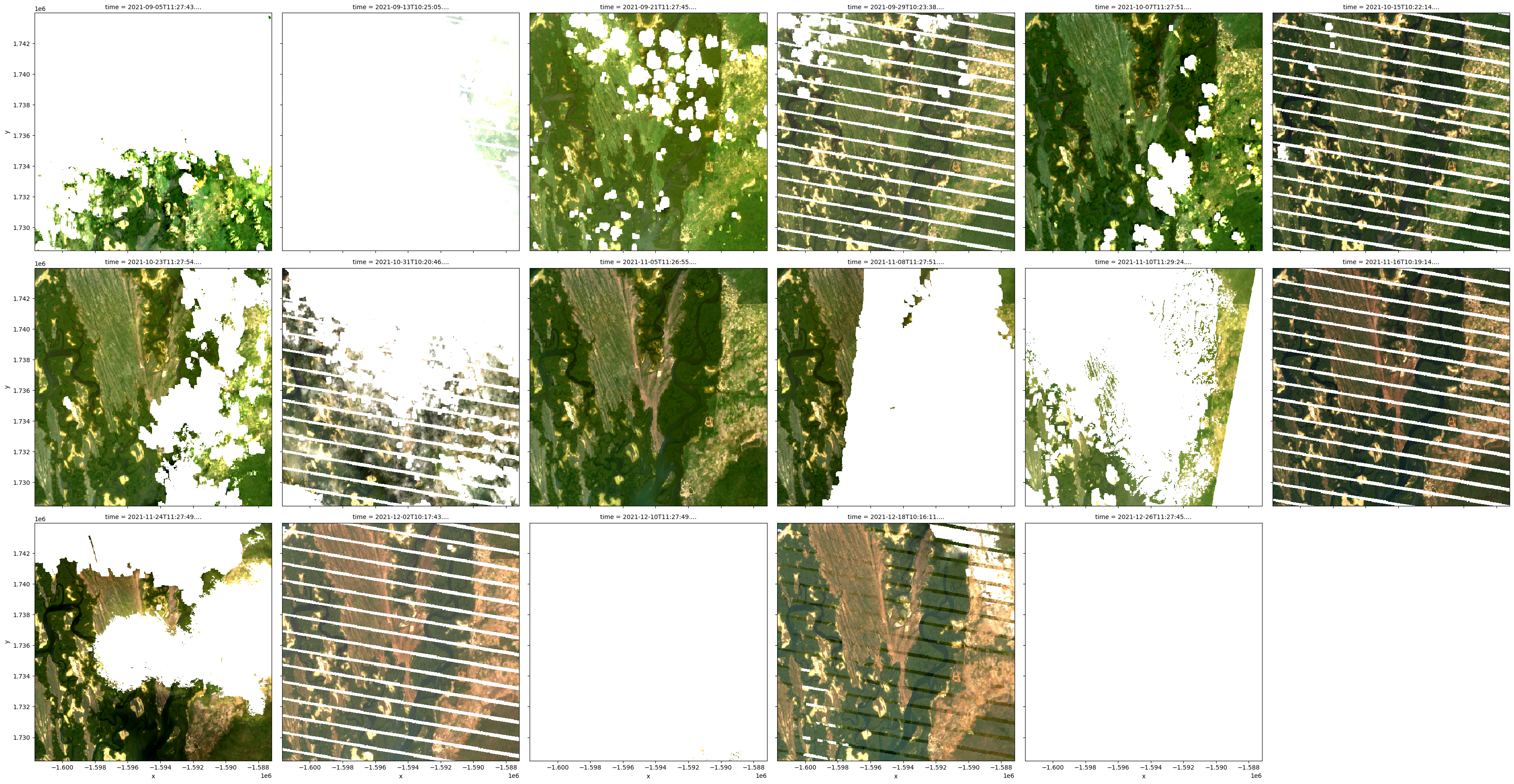
By default, load_ard applies a pixel quality mask to loaded data using the sensor’s pixel quality band.
For USGS Collection 2, the following mask parameters are applied to the qa_pixel band:
{
'cloud':"high_confidence",
'cloud_shadow':"high_confidence",
'nodata':True
}
For Sentinel 2, the following mask parameters are applied to the SCL band:
[
"cloud high probability",
"cloud medium probability",
"thin cirrus",
"cloud shadows",
"saturated or defective",
"no data"
]
Note: These masking parameters can be customised using the
categories_to_mask_lsparameter for USGS data, and thecategories_to_mask_s2parameter for Sentinel 2 data
We can de-activate pixel masking completely by setting mask_pixel_quality=False:
[7]:
# Load available data with cloud masking deactivated
ds = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr',
'ls9_sr'],
mask_pixel_quality=False,
resolution=(-30, 30),
**query)
# Plot single observation
rgb(ds, col='time', col_wrap=10)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
ls9_sr
Re-scaling Landsat C2 data
Loading 17 time steps
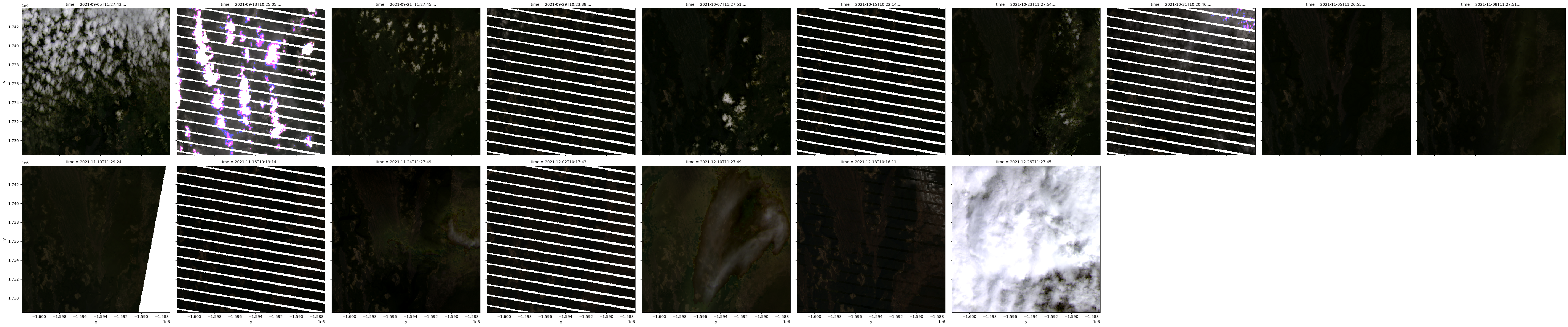
Filtering to non-cloudy observations¶
In addition to masking out cloud, load_ard allows you to discard any satellite observation that contains less than a minimum proportion of good quality (e.g. non-cloudy) pixels. This can be used to obtain a time series of only clear, cloud-free observations.
To discard all observations with less than X% good quality pixels, use the min_gooddata parameter. For example, min_gooddata=0.90 will return only observations where less than 10 % of pixels contain cloud, cloud shadow or other invalid data, resulting in a smaller number of clear, cloud free images being returned by the function:
[8]:
# Load available data filtered to 90% clear observations
ds = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr',
'ls9_sr'],
min_gooddata=0.90,
resolution=(-30, 30),
**query)
# Plot single observation
rgb(ds, index=2)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
ls9_sr
Counting good quality pixels for each time step
Filtering to 7 out of 17 time steps with at least 90.0% good quality pixels
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Loading 7 time steps
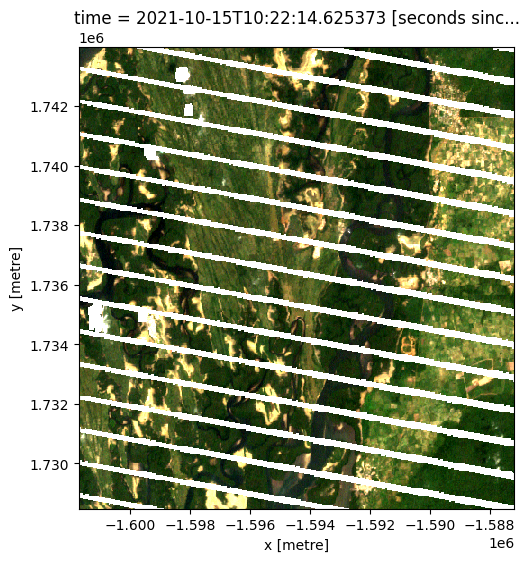
Applying morphological processing on the cloud mask¶
There are significant known limitations to the cloud masking algorithms employed by Sentinel-2 and Landsat. For example, bright objects like buildings and coastlines are commonly mistaken for cloud. We can improve on the false positives detected by Landsat and Sentinel-2’s pixel quality mask by applying binary moprhological image processing techniques (e.g.
binary_closing, binary_erosion etc.). The Open-Data-Cube library odc-algo has a function, odc.algo.mask_cleanup that can
perform a few of these operations. Below, we will try to improve the cloud mask by apply a number of these filters.
Feel free to experiment with the values in filters
[9]:
# set the filters to apply
filters = filters = [("opening", 4),("dilation", 2)]
# # Load data
data = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr',
'ls9_sr'],
resolution=(-30, 30),
mask_filters=filters,
**query)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
ls9_sr
Applying morphological filters to pq mask [('opening', 4), ('dilation', 2)]
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Loading 17 time steps
Below, you may notice that in the second and fourth images some of the false postives over the river/river banks have been removed by the morphological filters
[10]:
# plot the data
rgb(data, col="time", col_wrap=5)

Filtering data before load using a custom function¶
The load_ard function has a powerful predicate parameter that allows you to filter out satellite observations before they are actually loaded using a custom function. Some examples of where this may be useful include:
Filtering to return data from a specific season (e.g. summer, winter)
Filtering to return data acquired on a particular day of the year
Filtering to return data based on an external dataset (e.g. data acquired during specific climatic conditions such as drought or flood)
A predicate function should take a datacube.model.Dataset object as an input (e.g. as returned from dc.find_datasets(product='ls8_sr', **query)[0], and return either True or False. For example, a predicate function could be used to return True for only datasets acquired in April:
dataset.time.begin.month == 4
Filter to a single month¶
In the example below, we create a simple predicate function that will filter our data to return only satellite data acquired in April:
[11]:
def filter_april(dataset):
return dataset.time.begin.month == 4
# Load data that passes the `filter_april` function
ds = load_ard(dc=dc,
products=['ls5_sr',
'ls7_sr',
'ls8_sr'],
x=(-16.45, -16.6),
y=(13.675, 13.8),
time=('2016', '2018'),
measurements = ['red', 'green', 'blue'],
output_crs='epsg:6933',
predicate=filter_april,
resolution=(-30, 30),
group_by='solar_day')
# Print output data
print(ds)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
ls8_sr
Filtering datasets using filter function
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Loading 10 time steps
<xarray.Dataset>
Dimensions: (time: 10, y: 517, x: 484)
Coordinates:
* time (time) datetime64[ns] 2016-04-16T11:27:02.825796 ... 2018-04...
* y (y) float64 1.744e+06 1.744e+06 ... 1.729e+06 1.728e+06
* x (x) float64 -1.602e+06 -1.602e+06 ... -1.587e+06 -1.587e+06
spatial_ref int32 6933
Data variables:
red (time, y, x) float32 0.0824 0.08374 0.08391 ... 0.1524 0.15
green (time, y, x) float32 0.08861 0.08985 0.09015 ... 0.1053 0.1001
blue (time, y, x) float32 0.06472 0.06551 0.06568 ... 0.0665 0.06414
Attributes:
crs: epsg:6933
grid_mapping: spatial_ref
We can print the time steps returned by load_ard to verify that they now include only April observations (e.g. 2018-04-…):
[12]:
ds.time.values
[12]:
array(['2016-04-16T11:27:02.825796000', '2016-04-24T11:29:48.253992000',
'2017-04-03T11:27:01.738770000', '2017-04-11T11:29:47.043543000',
'2017-04-19T11:26:52.881313000', '2017-04-27T11:29:53.632066000',
'2018-04-06T11:26:52.293262000', '2018-04-14T11:28:13.576859000',
'2018-04-22T11:26:42.800959000', '2018-04-30T11:27:56.435231000'],
dtype='datetime64[ns]')
Filter to a single season¶
An example of a predicate function that will return data from a season of interest would look as follows:
def seasonal_filter(dataset, season=[12,1,2]):
#return true if month is in defined season
return dataset.time.begin.month in season
After applying this predicate function, running the following command demonstrates that our dataset only contains months during the Dec, Jan, Feb period
ds.time.dt.season :
<xarray.DataArray 'season' (time: 37)>
array(['DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF',
'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF',
'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF',
'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF', 'DJF',
'DJF'], dtype='<U3')
Coordinates:
* time (time) datetime64[ns] 2016-01-05T10:27:44.213284 ... 2017-12-26T10:23:43.129624
Loading Sentinel-2 data¶
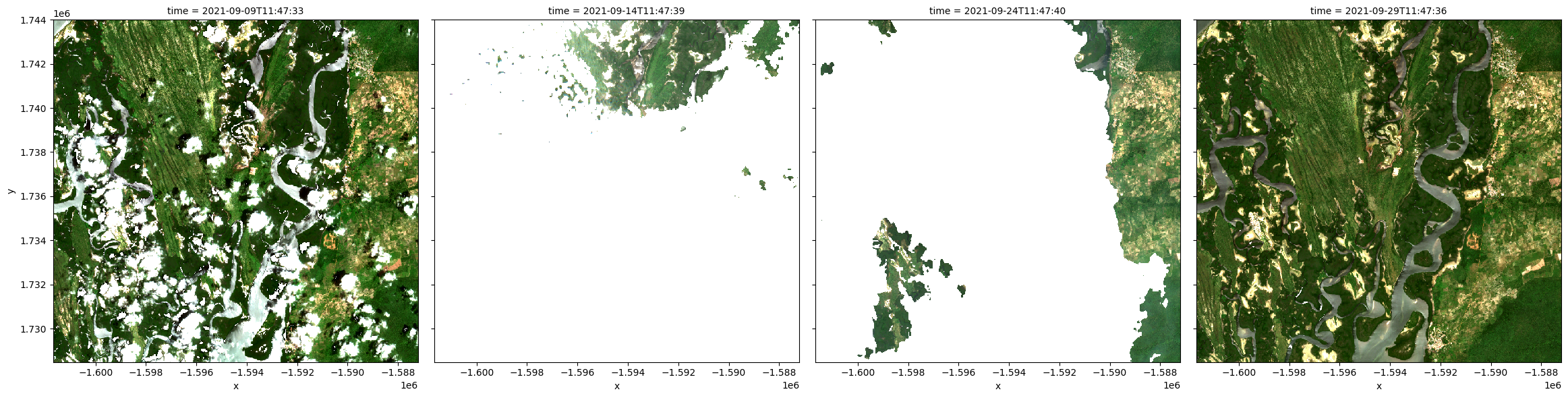
Data from the Sentinel-2A and Sentinel-2B satellites can also be loaded using load_ard. To do this, we need to specify the Sentinel-2 product (['s2a_l2a'] - both sensors S2a and S2b are covered by this product name) in place of the Landsat products above.
[13]:
# Load available data from S2
s2 = load_ard(dc=dc,
products=['s2_l2a'],
resolution=(-20, 20),
**query)
# # Print output data
print(s2)
Using pixel quality parameters for Sentinel 2
Finding datasets
s2_l2a
Applying pixel quality/cloud mask
Loading 24 time steps
<xarray.Dataset>
Dimensions: (time: 24, y: 776, x: 724)
Coordinates:
* time (time) datetime64[ns] 2021-09-04T11:47:39 ... 2021-12-28T11:...
* y (y) float64 1.744e+06 1.744e+06 ... 1.729e+06 1.728e+06
* x (x) float64 -1.602e+06 -1.602e+06 ... -1.587e+06 -1.587e+06
spatial_ref int32 6933
Data variables:
red (time, y, x) float32 1.062e+03 1.08e+03 ... 348.0 373.0
green (time, y, x) float32 1.184e+03 1.28e+03 ... 354.0 340.0
blue (time, y, x) float32 1.264e+03 1.346e+03 ... 119.0 136.0
Attributes:
crs: epsg:6933
grid_mapping: spatial_ref
Cloudy pixels are masked out by default from the resulting observations similarly to Landsat
[14]:
# Plot single observation
rgb(s2, index=[1,2,4,5])
Lazy loading with Dask¶
Rather than load data directly - which can take a long time and large amounts of memory - all datacube data can be lazy loaded using Dask. This can be a very useful approach for when you need to load large amounts of data without crashing your analysis, or if you want to subsequently scale your analysis by distributing tasks in parallel across multiple workers.
The load_ard function can be easily adapted to lazily load data rather than loading it into memory by providing a dask_chunks parameter using either the explicit or query syntax. The minimum required to lazily load data is dask_chunks={}, but chunking can also be performed spatially (e.g. dask_chunks={'x': 3000, 'y': 3000}) or by time (e.g. dask_chunks={'time': 1}) depending on the analysis being conducted. See the Dask
documentation for more information about setting chunk sizes.
To learn more about lazy-loading with Dask, see the Dask notebook.
[15]:
# Load available data from both S2 datasets
s2 = load_ard(dc=dc,
products=['s2_l2a'],
resolution=(-20, 20),
dask_chunks={'time':1,'x':500,'y':500},
**query)
# Print output data
print(s2)
Using pixel quality parameters for Sentinel 2
Finding datasets
s2_l2a
Applying pixel quality/cloud mask
Returning 24 time steps as a dask array
<xarray.Dataset>
Dimensions: (time: 24, y: 776, x: 724)
Coordinates:
* time (time) datetime64[ns] 2021-09-04T11:47:39 ... 2021-12-28T11:...
* y (y) float64 1.744e+06 1.744e+06 ... 1.729e+06 1.728e+06
* x (x) float64 -1.602e+06 -1.602e+06 ... -1.587e+06 -1.587e+06
spatial_ref int32 6933
Data variables:
red (time, y, x) float32 dask.array<chunksize=(1, 500, 500), meta=np.ndarray>
green (time, y, x) float32 dask.array<chunksize=(1, 500, 500), meta=np.ndarray>
blue (time, y, x) float32 dask.array<chunksize=(1, 500, 500), meta=np.ndarray>
Attributes:
crs: epsg:6933
grid_mapping: spatial_ref
Additional information¶
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Africa data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Slack channel or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
Github.
Compatible datacube version:
[16]:
print(datacube.__version__)
1.8.15
Last Tested:
[17]:
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
[17]:
'2023-08-14'