Combining satellite data with tidal modelling using OTPS¶
Keywords data used; landsat 5, data used; landsat 7, data used; landsat 8, tidal; modelling, intertidal, dask
Contexte¶
Ocean tides are the periodic rise and fall of the ocean caused by the gravitational pull of the moon and sun and the earth’s rotation. Tides in coastal areas can greatly influence how these environments appear in satellite imagery as water levels vary by up to 12 metres. To be able to study environmental processes along Africa’s coastline, it is vital to obtain data on tidal conditions at the exact moment each satellite image was acquired.
Description¶
This notebook demonstrates how to tidally tag remotely sensed imagery using functions from the deafrica_tools.coastal package so that images can be extracted or analysed by tidal stage (e.g. low, high, ebb, flow). These functions use the OTPS TPXO8 tidal model to calculate the height (relative to mean sea level) and stage of the tide at the exact moment each satellite image was acquired.
The notebook demonstrates how to:
Load an example time series of satellite data
Use the
tidal_tagfunction fromdeafrica_tools.coastalto model tide heights for each satellite observationUse tide height data to produce median composites of the coast at low and high tide
Swap a dataset’s dimensions to compute a rolling median along the
tide_heightdimensionCompute ebb or flow tide phase data to determine whether water levels were rising or falling in each satellite observation
Use the
tidal_statsfunction to evaluate any biases in the tidal conditions observed by a satellite
Getting started¶
To run this analysis, run all the cells in the notebook, starting with the « Load packages » cell.
Load packages¶
[1]:
%matplotlib inline
import datacube
import xarray as xr
import geopandas as gpd
import matplotlib.pyplot as plt
from datacube.utils.geometry import Geometry
from deafrica_tools.plotting import rgb, display_map
from deafrica_tools.datahandling import load_ard, mostcommon_crs
from deafrica_tools.coastal import tidal_tag, tidal_stats
from deafrica_tools.dask import create_local_dask_cluster
from deafrica_tools.areaofinterest import define_area
Set up a Dask cluster¶
Dask can be used to better manage memory use down and conduct the analysis in parallel. For an introduction to using Dask with Digital Earth Africa, see the Dask notebook.
Note: We recommend opening the Dask processing window to view the different computations that are being executed; to do this, see the Dask dashboard in DE Africa section of the Dask notebook.
To use Dask, set up the local computing cluster using the cell below.
[ ]:
create_local_dask_cluster()
Connect to the datacube¶
[3]:
dc = datacube.Datacube(app='Tidal_modelling')
Set up data query¶
First we set up a query to define the area, time period and other parameters required for loading data. In this example, we will load 30 years of Landsat 5, 7 and 8 data for the Geba River Estuary in southern Guinea-Bissau. We load the 'red', 'green', 'blue' bands so that we can plot the data as true colour imagery.
Note: The
dask_chunksparameter allows us to use Dask to lazily load data rather than load data directly into memory, as the standard loading method can take a long time and large amounts of memory. Lazy loading can be a very useful approach for when you need to load large amounts of data without crashing your analysis. In coastal applications, it allows us to load (using either.compute()or by plotting our data) only a small subset of observations from our entire time series (e.g. only low or high tide observations) without having to load the entire dataset into memory first, which can greatly decrease processing times.
Define location¶
To define the area of interest, there are two methods available:
By specifying the latitude, longitude, and buffer. This method requires you to input the central latitude, central longitude, and the buffer value in square degrees around the center point you want to analyze. For example,
lat = 10.338,lon = -1.055, andbuffer = 0.1will select an area with a radius of 0.1 square degrees around the point with coordinates (10.338, -1.055).By uploading a polygon as a
GeoJSON or Esri Shapefile. If you choose this option, you will need to upload the geojson or ESRI shapefile into the Sandbox using Upload Files button in the top left corner of the Jupyter Notebook interface. ESRI shapefiles must be uploaded with all the related files
in the top left corner of the Jupyter Notebook interface. ESRI shapefiles must be uploaded with all the related files (.cpg, .dbf, .shp, .shx). Once uploaded, you can use the shapefile or geojson to define the area of interest. Remember to update the code to call the file you have uploaded.
To use one of these methods, you can uncomment the relevant line of code and comment out the other one. To comment out a line, add the "#" symbol before the code you want to comment out. By default, the first option which defines the location using latitude, longitude, and buffer is being used.
[4]:
# Define the location
# Method 1: Specify the latitude, longitude, and buffer
aoi = define_area(lat=11.689, lon=-15.674, buffer=0.15)
# Method 2: Use a polygon as a GeoJSON or Esri Shapefile.
# aoi = define_area(vector_path='aoi.shp')
#Create a geopolygon and geodataframe of the area of interest
geopolygon = Geometry(aoi["features"][0]["geometry"], crs="epsg:4326")
geopolygon_gdf = gpd.GeoDataFrame(geometry=[geopolygon], crs=geopolygon.crs)
# Get the latitude and longitude range of the geopolygon
lat_range = (geopolygon_gdf.total_bounds[1], geopolygon_gdf.total_bounds[3])
lon_range = (geopolygon_gdf.total_bounds[0], geopolygon_gdf.total_bounds[2])
# Create a reusable query
query = {
'x': lon_range,
'y': lat_range,
'time': ('1988-01-01', '2018-12-31'),
'measurements': ['red', 'green', 'blue'],
'resolution': (-30, 30),
'dask_chunks': {}
}
We can preview the area that we will load data for:
[5]:
display_map(x=query['x'], y=query['y'])
[5]:
Load satellite time-series¶
To obtain some satellite data to analyse, we use the load_ard function to import a time series of Landsat 5, 7 and 8 observations as an xarray.Dataset. The input data does not need to be from Landsat: any remotely-sensed imagery with timestamps and spatial coordinates provide enough data to run the tidal model.
[6]:
# Identify the most common projection system in the input query
output_crs = mostcommon_crs(dc=dc, product='ls8_sr', query=query)
# Load available data from all three Landsat satellites
ds = load_ard(dc=dc,
products=['ls5_sr', 'ls7_sr', 'ls8_sr'],
output_crs=output_crs,
align=(15, 15),
ls7_slc_off=False,
group_by='solar_day',
**query)
# Print output data
print(ds)
Using pixel quality parameters for USGS Collection 2
Finding datasets
ls5_sr
ls7_sr
Ignoring SLC-off observations for ls7
ls8_sr
Applying pixel quality/cloud mask
Re-scaling Landsat C2 data
Returning 227 time steps as a dask array
<xarray.Dataset>
Dimensions: (time: 227, y: 1109, x: 1093)
Coordinates:
* time (time) datetime64[ns] 1988-07-17T10:52:47.658038 ... 2018-12...
* y (y) float64 1.309e+06 1.309e+06 ... 1.276e+06 1.276e+06
* x (x) float64 4.102e+05 4.102e+05 ... 4.429e+05 4.429e+05
spatial_ref int32 32628
Data variables:
red (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
green (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
blue (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
Attributes:
crs: epsg:32628
grid_mapping: spatial_ref
Model tide heights for each observation¶
We use the tidal_tag function from deafrica_coastaltools to associate each satellite observation in our timeseries with a tide height relative to mean sea level. This function uses the time and date of acquisition and the geographic location of each satellite observation as inputs to the OSU Tidal Prediction Software (OTPS) tidal model. From Sagar et al. 2015:
Note: The OTPS TPX08 tidal model consists of a multi-resolution bathymetric grid solution, with a 1/6° solution in the global open ocean, and a 1/30° local resolution solution to improve modelling in complex shallow water environments. The OTPS model is based on a system of linear partial differential equations, called Laplace’s tidal equations, parametrised with nine harmonic tidal constituents. The model is fitted to track-averaged TOPEX/Poseidon altimeter data collected from 1992 to 2016 and Jason-1 (Poseidon 2) altimeter data from 2002 to 2013, enabling estimation of the tidal height and harmonic constituents at discrete temporal epochs and spatial locations.
[7]:
# Model tide heights
ds_tidal = tidal_tag(ds)
# Print output data
print(ds_tidal)
Setting tide modelling location from dataset centroid: -15.67, 11.69
<xarray.Dataset>
Dimensions: (time: 227, y: 1109, x: 1093)
Coordinates:
* time (time) datetime64[ns] 1988-07-17T10:52:47.658038 ... 2018-12...
* y (y) float64 1.309e+06 1.309e+06 ... 1.276e+06 1.276e+06
* x (x) float64 4.102e+05 4.102e+05 ... 4.429e+05 4.429e+05
spatial_ref int32 32628
Data variables:
red (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
green (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
blue (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
tide_height (time) float64 -0.194 -0.605 0.168 ... 0.535 -0.217 -0.821
Attributes:
crs: epsg:32628
grid_mapping: spatial_ref
The function will automatically select a tide modelling location based on the dataset centroid. It will then output modelled tide heights as a new tide_height variable in the xarray.Dataset (the variable should appear under Data variables above).
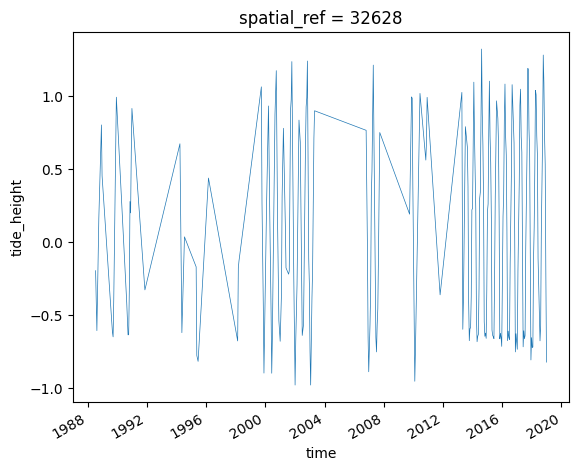
We can easily plot this new variable to inspect the range of tide heights observed by the satellites in our timeseries. In this example, our observed tide heights range from approximately -1.0 to 1.0 m relative to Mean Sea Level:
[8]:
ds_tidal.tide_height.plot(linewidth=0.5)
[8]:
[<matplotlib.lines.Line2D at 0x7f4bca0ea560>]
Example tide height analysis¶
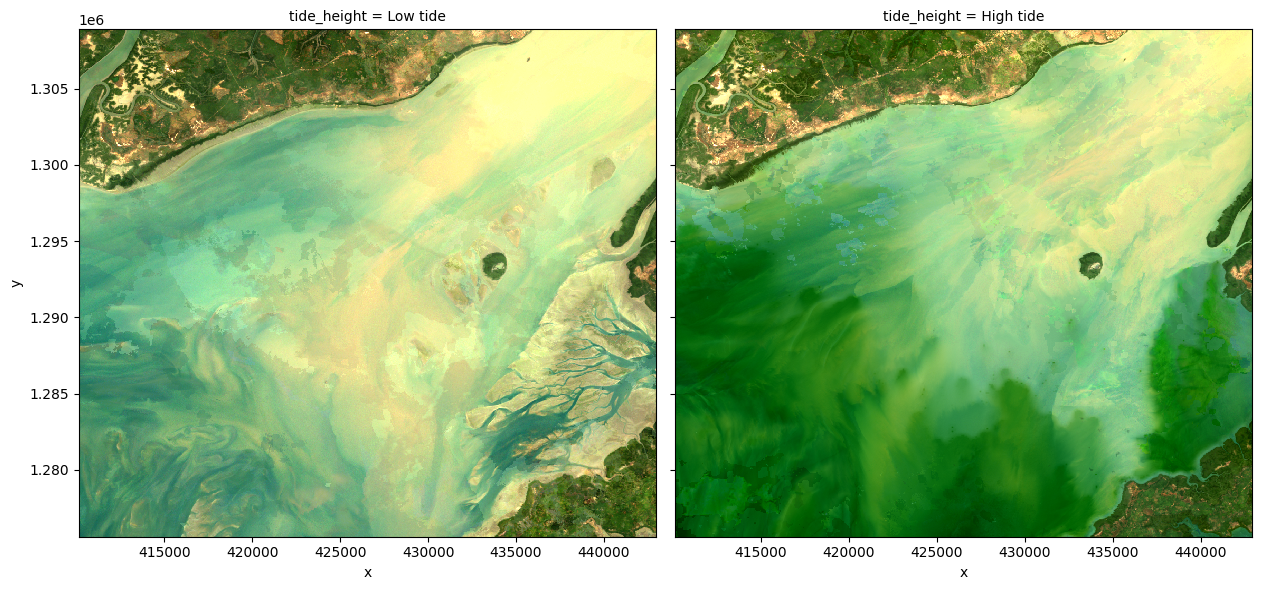
To demonstrate how tidally tagged images can be used to produce composites of high and low tide imagery, we can compute the lowest 5% and highest 5% percent of tide heights, and use these to filter our observations. We can then combine and plot these filtered observations to visualise what the landscape looks like at low and high tide:
Note: We recommend opening the Dask processing window to view the different computations that are being executed; to do this, see the Dask dashboard in DE Africa section of the Dask notebook.
Note: An alternative approach to combining observations into a composite can be achieved using a geomedian, which ensures band relationships are kept consistent. More information is provided in the Generating geomedian composites notebook.
[9]:
# Calculate the lowest and highest 5% of tides
lowest_5, highest_5 = ds_tidal.tide_height.quantile([0.05, 0.95]).values
# Filter our data to low and high tide observations
filtered_low = ds_tidal.where(ds_tidal.tide_height <= lowest_5, drop=True)
filtered_high = ds_tidal.where(ds_tidal.tide_height >= highest_5, drop=True)
# Take the simple median of each set of low and high tide observations to
# produce a composite (alternatively, observations could be combined
# using a geomedian to keep band relationships consistent)
median_low = filtered_low.median(dim='time', keep_attrs=True)
median_high = filtered_high.median(dim='time', keep_attrs=True)
# Combine low and high tide medians into a single dataset and give
# each layer a meaningful name
ds_highlow = xr.concat([median_low, median_high], dim='tide_height')
ds_highlow['tide_height'] = ['Low tide', 'High tide']
# Plot low and high tide medians side-by-side
rgb(ds_highlow, col='tide_height')
Swapping dimensions¶
The tidal_tag function allows you to use tide heights as the primary dimension in the dataset, rather than time. Setting swap_dims=True will swap the time dimension in the original xarray.Dataset to the new tide_height variable.
[10]:
# Model tide heights
ds_tidal = tidal_tag(ds, swap_dims=True)
# Print output data
print(ds_tidal)
Setting tide modelling location from dataset centroid: -15.67, 11.69
<xarray.Dataset>
Dimensions: (tide_height: 227, y: 1109, x: 1093)
Coordinates:
* y (y) float64 1.309e+06 1.309e+06 ... 1.276e+06 1.276e+06
* x (x) float64 4.102e+05 4.102e+05 ... 4.429e+05 4.429e+05
spatial_ref int32 32628
* tide_height (tide_height) float64 -0.978 -0.978 -0.952 ... 1.284 1.325
Data variables:
red (tide_height, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
green (tide_height, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
blue (tide_height, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
Attributes:
crs: epsg:32628
grid_mapping: spatial_ref
The dataset now contains three dimensions: tide_height, x and y.
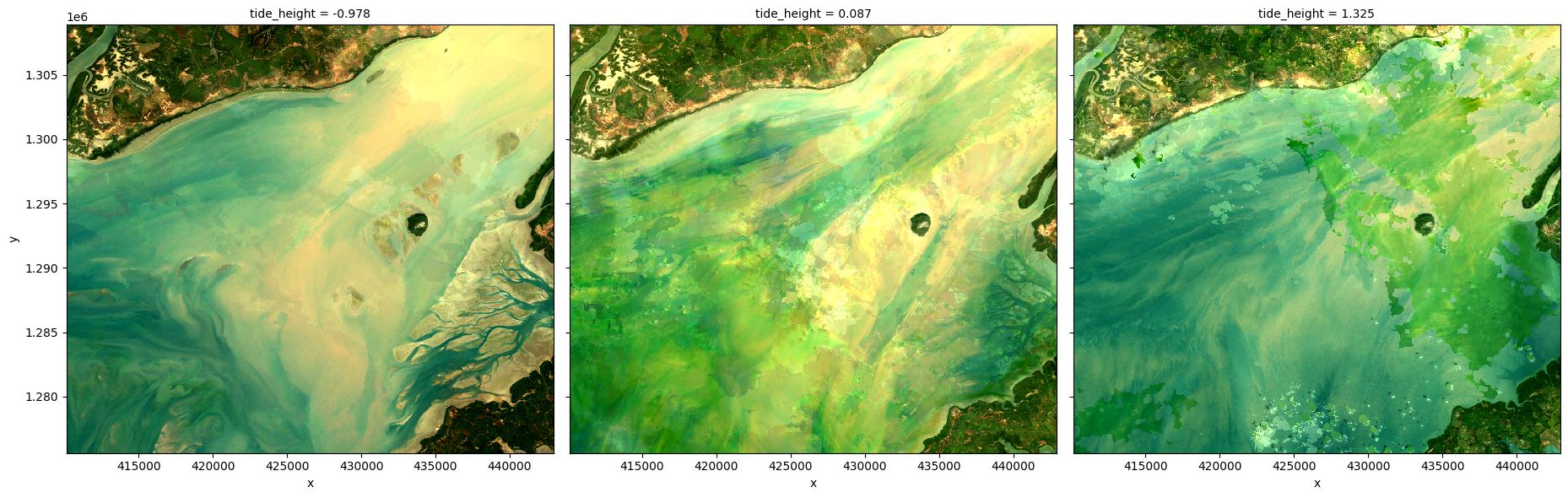
This can make it easier to analyse the data with respect to tide, e.g. computing a rolling median by tide height (e.g. along the tide_height dimension), as is done in the following cell.
Note: We recommend opening the Dask processing window to view the different computations that are being executed; to do this, see the Dask dashboard in DE Africa section of the Dask notebook.
[11]:
# First we need to update the chunks used by Dask to allow us to do a
# rolling median without having to load all our data into memory first
ds_rechunked = ds_tidal.chunk(chunks={'tide_height': 13})
# Compute a rolling median that will go through every satellite
# observation, and take the median of that timestep and its 15 neighbours
ds_rolling = ds_rechunked.rolling(tide_height=13,
center=True,
min_periods=1).median()
# Plot the lowest, median and highest tide rolling median image
rgb(ds_rolling, index_dim='tide_height', index=[0, len(ds_tidal.tide_height)//2, -1])
Modelling ebb and flow tidal phases¶
The tidal_tag function also allows us to determine whether each satellite observation was taken while the tide was rising/incoming (flow tide) or falling/outgoing (ebb tide) by setting ebb_flow=True. This is achieved by comparing tide heights 15 minutes before the before and after the observed satellite observation.
Ebb and flow data can provide valuable contextual information for interpreting satellite imagery, particularly in tidal flat or mangrove forest environments where water may remain in the landscape for considerable time after the tidal peak.
[12]:
# Model tide heights
ds_tidal = tidal_tag(ds, ebb_flow=True)
# Print output data
print(ds_tidal)
Setting tide modelling location from dataset centroid: -15.67, 11.69
Modelling tidal phase (e.g. ebb or flow)
<xarray.Dataset>
Dimensions: (time: 227, y: 1109, x: 1093)
Coordinates:
* time (time) datetime64[ns] 1988-07-17T10:52:47.658038 ... 2018-12...
* y (y) float64 1.309e+06 1.309e+06 ... 1.276e+06 1.276e+06
* x (x) float64 4.102e+05 4.102e+05 ... 4.429e+05 4.429e+05
spatial_ref int32 32628
Data variables:
red (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
green (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
blue (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
tide_height (time) float64 -0.194 -0.605 0.168 ... 0.535 -0.217 -0.821
ebb_flow (time) <U4 'Flow' 'Flow' 'Ebb' 'Ebb' ... 'Flow' 'Flow' 'Flow'
Attributes:
crs: epsg:32628
grid_mapping: spatial_ref
We now have data giving us the both the tide height and tidal phase (“ebb” or “flow”) for every satellite image:
[13]:
ds_tidal[['time', 'tide_height', 'ebb_flow']].to_dataframe()
[13]:
| tide_height | ebb_flow | spatial_ref | |
|---|---|---|---|
| time | |||
| 1988-07-17 10:52:47.658038 | -0.194 | Flow | 32628 |
| 1988-08-18 10:52:49.179013 | -0.605 | Flow | 32628 |
| 1988-10-05 10:52:43.680006 | 0.168 | Ebb | 32628 |
| 1988-10-21 10:52:37.662000 | 0.334 | Ebb | 32628 |
| 1988-12-08 10:52:31.815006 | 0.805 | Flow | 32628 |
| ... | ... | ... | ... |
| 2018-10-24 11:21:58.388427 | 1.094 | Flow | 32628 |
| 2018-11-09 11:22:01.151598 | 0.730 | Flow | 32628 |
| 2018-11-25 11:22:00.936674 | 0.535 | Flow | 32628 |
| 2018-12-11 11:21:57.773631 | -0.217 | Flow | 32628 |
| 2018-12-27 11:21:57.806437 | -0.821 | Flow | 32628 |
227 rows × 3 columns
We could for example use this data to filter our observations to keep ebbing phase observations only:
[14]:
ds_tidal.where(ds_tidal.ebb_flow == 'Ebb', drop=True)
[14]:
<xarray.Dataset>
Dimensions: (time: 106, y: 1109, x: 1093)
Coordinates:
* time (time) datetime64[ns] 1988-10-05T10:52:43.680006 ... 2018-10...
* y (y) float64 1.309e+06 1.309e+06 ... 1.276e+06 1.276e+06
* x (x) float64 4.102e+05 4.102e+05 ... 4.429e+05 4.429e+05
spatial_ref int32 32628
Data variables:
red (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
green (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
blue (time, y, x) float32 dask.array<chunksize=(1, 1109, 1093), meta=np.ndarray>
tide_height (time) float64 0.168 0.334 -0.648 -0.395 ... 0.065 0.724 1.284
ebb_flow (time) object 'Ebb' 'Ebb' 'Ebb' 'Ebb' ... 'Ebb' 'Ebb' 'Ebb'
Attributes:
crs: epsg:32628
grid_mapping: spatial_refEvaluating observed vs. all modelled tide heights¶
The complex behaviour of tides mean that a sun synchronous sensor like Landsat does not observe the full range of the tidal cycle at all locations. Biases in the proportion of the tidal range observed by satellites can prevent us from obtaining data on areas of the coastline exposed or inundated at the extremes of the tidal range. This can risk gaining misleading insights into the true extent of the area of the coastline affected by tides, and make it difficult to compare high or low tide images fairly in different locations.
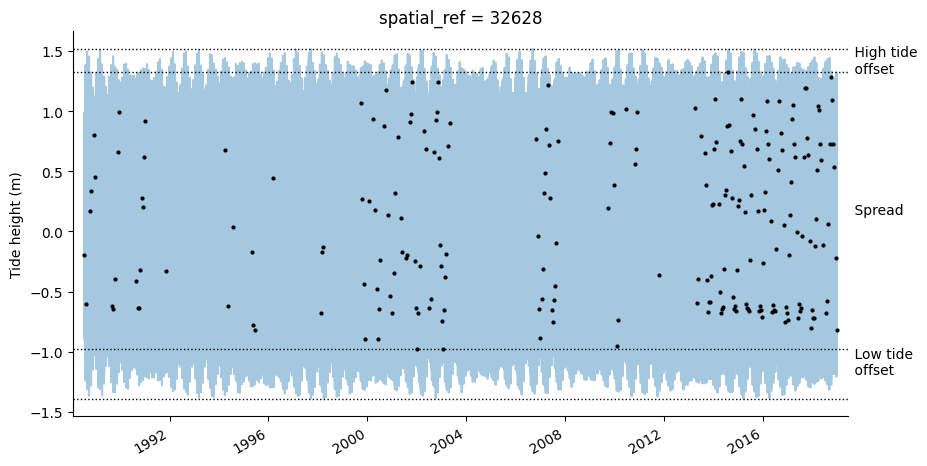
The tidal_stats function can assist in evaluating how the range of tides observed by satellites compare to the full tidal range. It works by using the OTPS tidal model to model tide heights at a regular interval (every two hours by default) across the entire time period covered by the input satelliter timeseries dataset. This is then compared against the tide heights in observed by the satellite and used to calculate a range of statistics and a plot that summarises potential biases in the
data.
Note: For a more detailed discussion of the issue of tidal bias in sun-synchronous satellite observations of the coastline, refer to the “Limitations and future work” section in Bishop-Taylor et al. 2018.
[15]:
out_stats = tidal_stats(ds)
Setting tide modelling location from dataset centroid: -15.67, 11.69
79% of the full 2.91 m modelled tidal range is observed at this location.
The lowest 14% and highest 7% of tides are never observed.
Observed tides do not increase or decrease significantly over the ~31 year period.
All tides do not increase or decrease significantly over the ~31 year period.
The function also outputs a pandas.Series object containing a set of statistics that compare the observed vs. full modelled tidal ranges. These statistics include:
tidepost_lat: latitude used for modelling tide heights
tidepost_lon: longitude used for modelling tide heights
observed_min_m: minimum tide height observed by the satellite (in metre units)
all_min_m: minimum tide height from full modelled tidal range (in metre units)
observed_max_m: maximum tide height observed by the satellite (in metre units)
all_max_m: maximum tide height from full modelled tidal range (in metre units)
observed_range_m: tidal range observed by the satellite (in metre units)
all_range_m: full modelled tidal range (in metre units)
spread_m: proportion of the full modelled tidal range observed by the satellite (see Bishop-Taylor et al. 2018)
low_tide_offset: proportion of the lowest tides never observed by the satellite (see Bishop-Taylor et al. 2018)
high_tide_offset: proportion of the highest tides never observed by the satellite (see Bishop-Taylor et al. 2018)
observed_slope: slope of any relationship between observed tide heights and time
all_slope: slope of any relationship between all modelled tide heights and time
observed_pval: significance/p-value of any relationship between observed tide heights and time
all_pval: significance/p-value of any relationship between all modelled tide heights and time
[16]:
print(out_stats)
tidepost_lat 11.689
tidepost_lon -15.674
observed_mean_m 0.086
all_mean_m 0.000
observed_min_m -0.978
all_min_m -1.393
observed_max_m 1.325
all_max_m 1.518
observed_range_m 2.303
all_range_m 2.911
spread 0.791
low_tide_offset 0.143
high_tide_offset 0.066
observed_slope 0.005
all_slope -0.000
observed_pval 0.287
all_pval 0.370
dtype: float64
Additional information¶
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Africa data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Slack channel or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
Github.
Compatible datacube version:
[17]:
print(datacube.__version__)
1.8.15
Last Tested:
[18]:
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
[18]:
'2023-08-14'