Generating animated time series using xr_animation¶
Products used: s2_l2a
Keywords data used; sentinel-2, data methods; animation, band index; NDWI
Contexte¶
Animations can be a powerful method for visualising change in the landscape across time using satellite imagery. Satellite data from Digital Earth Africa is an ideal subject for animations as it has been georeferenced, processed to analysis-ready surface reflectance, and stacked into a spatio-temporal “data cube”, allowing landscape conditions to be extracted and visualised consistently across time.
Using the DE Africa functions in Scripts/deafrica_plotting, which are based on matplotlib.animation and xarray, we can take a time series of Digital Earth Africa satellite imagery and export a visually appealing time series animation that shows how any location in Africa has changed.
Description¶
This notebook demonstrates how to:
Import a time series of cloud-free satellite imagery from multiple satellites (i.e. Sentinel-2A and -2B) as an xarray dataset
Plot the data as a three band time series animation
Plot the data as a one band time series animation
Export the resulting animations as either a GIF or MP4 file
Add custom vector overlays
Apply custom image processing functions to each animation frame
Getting started¶
To run this analysis, run all the cells in the notebook, starting with the « Load packages » cell.
Load packages¶
[1]:
%matplotlib inline
# Force GeoPandas to use Shapely instead of PyGEOS
# In a future release, GeoPandas will switch to using Shapely by default.
import os
os.environ['USE_PYGEOS'] = '0'
import datacube
import skimage.exposure
import geopandas as gpd
import matplotlib.pyplot as plt
from IPython.display import Image
from datacube.utils.geometry import Geometry
from deafrica_tools.plotting import xr_animation, rgb
from deafrica_tools.datahandling import load_ard
from deafrica_tools.areaofinterest import define_area
Connect to the datacube¶
[2]:
dc = datacube.Datacube(app='Animated_timeseries')
Select location¶
To define the area of interest, there are two methods available:
By specifying the latitude, longitude, and buffer. This method requires you to input the central latitude, central longitude, and the buffer value in square degrees around the center point you want to analyze. For example,
lat = 10.338,lon = -1.055, andbuffer = 0.1will select an area with a radius of 0.1 square degrees around the point with coordinates (10.338, -1.055).By uploading a polygon as a
GeoJSON or Esri Shapefile. If you choose this option, you will need to upload the geojson or ESRI shapefile into the Sandbox using Upload Files button in the top left corner of the Jupyter Notebook interface. ESRI shapefiles must be uploaded with all the related files
in the top left corner of the Jupyter Notebook interface. ESRI shapefiles must be uploaded with all the related files (.cpg, .dbf, .shp, .shx). Once uploaded, you can use the shapefile or geojson to define the area of interest. Remember to update the code to call the file you have uploaded.
To use one of these methods, you can uncomment the relevant line of code and comment out the other one. To comment out a line, add the "#" symbol before the code you want to comment out. By default, the first option which defines the location using latitude, longitude, and buffer is being used.
If running the notebook for the first time, keep the default settings below. This will demonstrate how the analysis works and provide meaningful results.
[3]:
# Method 1: Specify the latitude, longitude, and buffer
aoi = define_area(lat=8.57, lon=-2.4, buffer=0.05)
# Method 2: Use a polygon as a GeoJSON or Esri Shapefile.
# aoi = define_area(vector_path='aoi.shp')
#Create a geopolygon and geodataframe of the area of interest
geopolygon = Geometry(aoi["features"][0]["geometry"], crs="epsg:4326")
geopolygon_gdf = gpd.GeoDataFrame(geometry=[geopolygon], crs=geopolygon.crs)
# Get the latitude and longitude range of the geopolygon
lat_range = (geopolygon_gdf.total_bounds[1], geopolygon_gdf.total_bounds[3])
lon_range = (geopolygon_gdf.total_bounds[0], geopolygon_gdf.total_bounds[2])
Load satellite data from datacube¶
We can use the load_ard function to load data from multiple satellites (i.e. Sentinel-2A and -2B), and return a single xarray.Dataset containing only observations with a minimum percentage of good quality pixels. This will allow us to create a visually appealing time series animation of observations that are not affected by cloud.
In the example below, we request that the function returns only observations which are 95% free of clouds and other poor quality pixels by specifyinge min_gooddata=0.95.
[4]:
# Create a reusable query
query = {
'x': lon_range,
'y': lat_range,
'time': ('2017-10-10', '2018-01-10'),
'resolution': (-20, 20)
}
[5]:
# Load available data
ds = load_ard(dc=dc,
products=['s2_l2a'],
measurements=['red', 'green', 'blue', 'nir_1', 'swir_1', 'swir_2'],
group_by='solar_day',
output_crs='epsg:32630',
min_gooddata=0.95,
mask_pixel_quality=True,
**query)
# Print output data
print(ds)
Using pixel quality parameters for Sentinel 2
Finding datasets
s2_l2a
Counting good quality pixels for each time step
Filtering to 13 out of 19 time steps with at least 95.0% good quality pixels
Applying pixel quality/cloud mask
Loading 13 time steps
<xarray.Dataset>
Dimensions: (time: 13, y: 555, x: 552)
Coordinates:
* time (time) datetime64[ns] 2017-10-17T10:40:55 ... 2018-01-10T10:...
* y (y) float64 9.529e+05 9.529e+05 ... 9.418e+05 9.418e+05
* x (x) float64 5.605e+05 5.605e+05 ... 5.715e+05 5.715e+05
spatial_ref int32 32630
Data variables:
red (time, y, x) float32 440.0 447.0 465.0 ... 526.0 537.0 557.0
green (time, y, x) float32 677.0 682.0 690.0 ... 683.0 679.0 668.0
blue (time, y, x) float32 250.0 274.0 285.0 ... 547.0 548.0 540.0
nir_1 (time, y, x) float32 3.098e+03 3.123e+03 ... 446.0 626.0
swir_1 (time, y, x) float32 1.671e+03 1.696e+03 ... 399.0 442.0
swir_2 (time, y, x) float32 783.0 775.0 773.0 ... 307.0 311.0 340.0
Attributes:
crs: epsg:32630
grid_mapping: spatial_ref
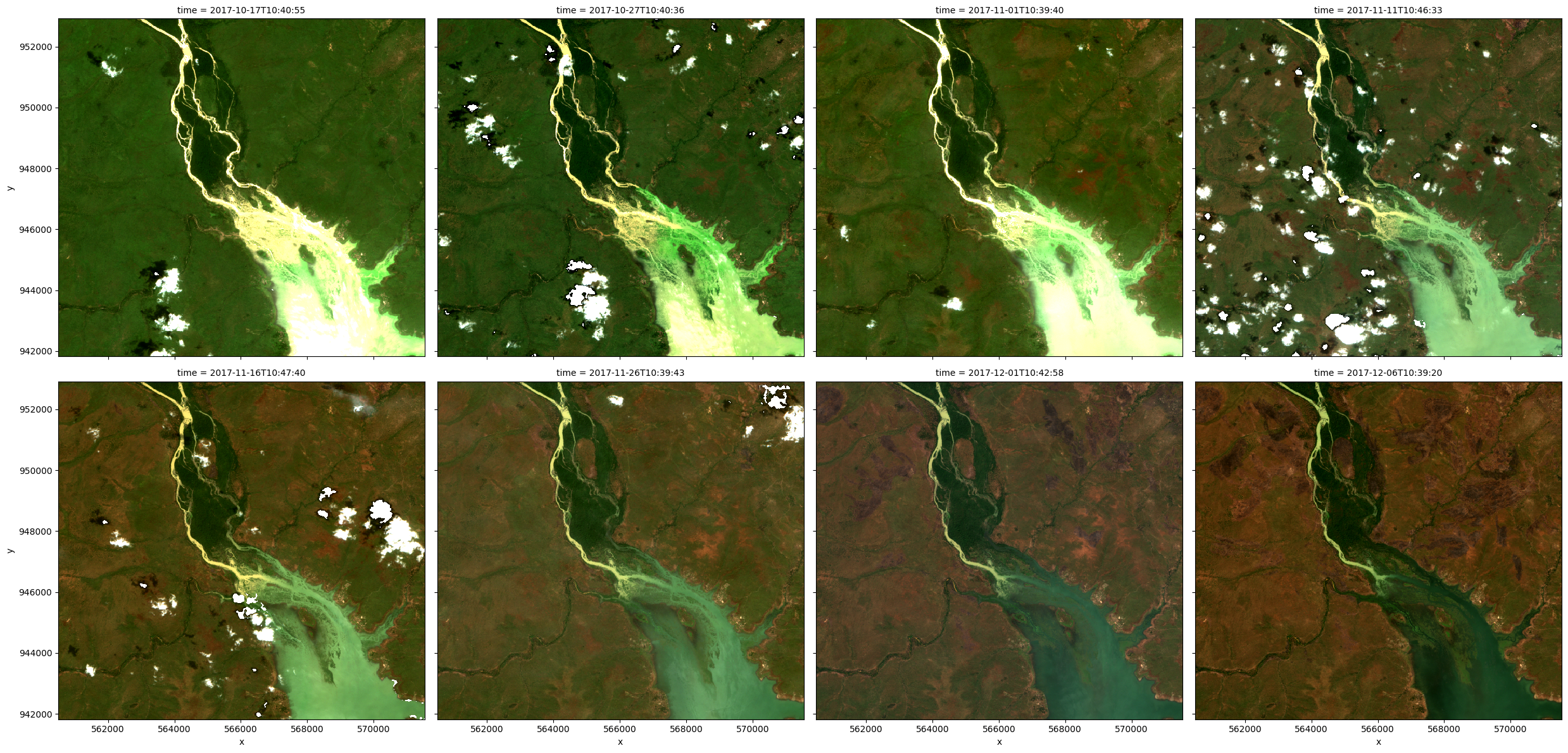
To get a quick idea of what the data looks like, we can plot the dataset in true colour using the rgb function:
[6]:
# Plot images from the dataset
rgb(ds, index=list(range(0, 8)))
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
Plot time series as a RGB/three band animated GIF¶
The xr_animation() function is based on functionality within matplotlib.animation. It takes an xarray.Dataset and exports a one band or three band (e.g. true or false colour) GIF or MP4 animation showing changes in the landscape across time.
['red', 'green', 'blue'] satellite bands to generate a true colour RGB animation.interval, and the width of the animation to 300 pixels using width_pixels. If these parameters are not specified, the function will use a default interval value of 100 milliseconds, and a default width_pixels value of 500 pixels.[7]:
# Produce time series animation of red, green and blue bands
xr_animation(ds=ds,
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.gif',
interval=200,
width_pixels=300)
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
[7]:
<IPython.core.display.Image object>
We can also use different band combinations (e.g. false colour) using bands, add additional text using show_text, and change the font size using annotation_kwargs, which passes a dictionary of values to the matplotlib plt.annotate function (see matplotlib.pyplot.annotate for options).
The function will automatically select an appropriate colour stretch by clipping the data to remove outliers/extreme values smaller or greater than the 2 and 98th percentiles (e.g. similar to xarray’s robust=True). This can be controlled further with the percentile_stretch parameter. For example, setting percentile_stretch=(0.01, 0.99) will apply a colour stretch with less contrast:
[8]:
# Produce time series animation of red, green and blue bands
xr_animation(ds=ds,
bands=['swir_2', 'nir_1', 'green'],
output_path='animated_timeseries.gif',
width_pixels=300,
show_text='Time-series animation',
percentile_stretch=(0.01, 0.99),
annotation_kwargs={'fontsize': 25})
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
[8]:
<IPython.core.display.Image object>
Plotting single band animations¶
It is also possible to plot a single band image instead of a three band image. For example, we could plot an index like the Normalized Difference Water Index (NDWI), which has high values where a pixel is likely to be open water (e.g. NDWI > 0).
By default the colour bar limits are set based on percentile_stretch which will discard outliers/extreme values to optimise the colour stretch (set percentile_stretch=(0.0, 1.00) to show the full range of values from min to max).
[9]:
# Compute NDWI using the formula (green - nir) / (green + nir).
# This will calculate NDWI for every time-step in the dataset:
ds['NDWI'] = ((ds.green - ds.nir_1) /
(ds.green + ds.nir_1))
# Produce time series animation of NDWI:
xr_animation(ds=ds,
output_path='animated_timeseries.gif',
bands=['NDWI'],
show_text='NDWI',
width_pixels=300)
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
[9]:
<IPython.core.display.Image object>
We can customise animations based on a single band like NDWI by specifying parameters using imshow_kwargs, which is passed to the matplotlib plt.imshow function (see link for options). For example, we can use a more appropriate blue colour scheme with 'cmap': 'Blues', and set 'vmin': 0.0, 'vmax': 0.5 to overrule the default colour bar limits with manually specified values:
[10]:
# Produce time series animation using a custom colour scheme and limits:
xr_animation(ds=ds,
bands='NDWI',
output_path='animated_timeseries.gif',
width_pixels=300,
show_text='NDWI',
imshow_kwargs={'cmap': 'Blues', 'vmin': 0.0, 'vmax': 0.5},
colorbar_kwargs={'colors': 'black'})
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
[10]:
<IPython.core.display.Image object>
One band animations show a colour bar by default, but this can be disabled using show_colorbar:
[11]:
# Produce time series animation using a custom colour scheme and limits:
xr_animation(ds=ds,
bands='NDWI',
output_path='animated_timeseries.gif',
width_pixels=300,
show_text='NDWI',
show_colorbar=False,
imshow_kwargs={'cmap': 'Blues', 'vmin': 0.0, 'vmax': 0.5})
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
[11]:
<IPython.core.display.Image object>
One band animations show a colour bar by default, but this can be disabled:
Available output formats¶
The above examples have focused on exporting animated GIFs, but MP4 files can also be generated. The two formats have their own advantages and disadvantages:
.mp4: fast to generate, smallest file sizes and highest quality; suitable for Twitter/social media and recent versions of Powerpoint.gif: slow to generate, large file sizes, low rendering quality; suitable for all versions of Powerpoint and Twitter/social media
Note: To preview a
.mp4file from within JupyterLab, find the file (e.g. “animated_timeseries.mp4”) in the file browser on the left, right click, and select “Open in New Browser Tab”.
[12]:
# Animate datasets as a MP4 file
xr_animation(ds=ds,
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.mp4')
Exporting animation to animated_timeseries.mp4
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)

Adding vector overlays¶
The animation code supports plotting vector files (e.g. ESRI Shapefiles or GeoJSON) over the top of satellite imagery. To do this, we first load the file using geopandas, and pass this to xr_animation using the show_gdf parameter:
[13]:
# Get shapefile path
poly_gdf = gpd.read_file('../Supplementary_data/Animated_timeseries/example_polygon.shp')
# Produce time series animation
xr_animation(ds=ds,
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.gif',
width_pixels=300,
show_gdf=poly_gdf)
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
[13]:
<IPython.core.display.Image object>
You can customise styling for vector overlays by including a column called 'color' in the geopandas.GeoDataFrame object:
[14]:
# Assign a colour to the GeoDataFrame
poly_gdf['color'] = 'red'
# Produce time series animation
xr_animation(ds=ds,
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.gif',
width_pixels=300,
show_gdf=poly_gdf)
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
[14]:
<IPython.core.display.Image object>
Plotting vectors by time¶
It can be useful to plot vector features over the top of imagery at specific times in an animation. For example, in this example we may want to plot different vectors depending on how full our waterbody was during our analysis period.
To do this, we can create new start_time and end_time columns in our geopandas.GeoDataFrame that tell xr_animation how long to plot each feature over the top of our imagery:
Note: Dates can be provided in any string format that can be converted using the pandas.to_datetime() function. For example, “2009”, “2009-11”, “2009-11-01” etc.
[15]:
# Assign start and end times to each feature
poly_gdf['start_time'] = ['2017-09', '2017-11', '2017-12', '2018-01']
poly_gdf['end_time'] = ['2017-11', '2017-12', '2018-01', '2018-02']
# Preview the updated geopandas.GeoDataFrame
poly_gdf
[15]:
| FID | geometry | color | start_time | end_time | |
|---|---|---|---|---|---|
| 0 | 0 | MULTIPOLYGON (((565920.000 946110.000, 565950.... | red | 2017-09 | 2017-11 |
| 1 | 1 | MULTIPOLYGON (((567300.000 945180.000, 567300.... | red | 2017-11 | 2017-12 |
| 2 | 2 | MULTIPOLYGON (((568830.000 944130.000, 568800.... | red | 2017-12 | 2018-01 |
| 3 | 3 | MULTIPOLYGON (((568830.000 944130.000, 568800.... | red | 2018-01 | 2018-02 |
We can now pass the updated geopandas.GeoDataFrame to xr_animation. We will also change our vector colours to a semi-transperant blue, and give them a blue outline using the gdf_kwargs parameter:
[16]:
# Set vector features to a semi-transperant blue to represent water
poly_gdf['color'] = '#0066ff90'
# Produce time series animation
xr_animation(ds=ds,
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.gif',
width_pixels=300,
show_gdf=poly_gdf,
gdf_kwargs={'edgecolor': 'blue'})
# Plot animated gif
plt.close()
Image(filename='animated_timeseries.gif')
Exporting animation to animated_timeseries.gif
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
/usr/local/lib/python3.10/dist-packages/matplotlib/cm.py:478: RuntimeWarning: invalid value encountered in cast
xx = (xx * 255).astype(np.uint8)
[16]:
<IPython.core.display.Image object>
Custom image processing functions¶
The image_proc_funcs parameter allows you to pass custom image processing functions that will be applied to each frame of your animation as they are rendered. This can be a powerful way to produce visually appealing animations - some example applications include:
Improving brightness, saturation or contrast
Sharpening your images
Histogram matching or equalisation
To demonstrate this, we will apply two functions from the skimage.exposure module which contains many powerful image processing algorithms:
skimage.exposure.rescale_intensitywill first scale our data between 0.0 and 1.0 (required for step 2)skimage.exposure.equalize_adapthistwill take this re-scaled data and apply an alogorithm that will enhance and contrast local details of the image
Note: Functions supplied to
image_proc_funcsare applied one after another to each timestep inds(e.g. each frame in the animation). Any custom function can be supplied toimage_proc_funcs, providing it both accepts and outputs anumpy.ndarraywith a shape of(y, x, bands).
[17]:
# Aniamte data after applying custom image processing functions to each frame
xr_animation(ds=ds.fillna(0),
bands=['red', 'green', 'blue'],
output_path='animated_timeseries.gif',
width_pixels=300,
image_proc_funcs=[skimage.exposure.rescale_intensity,
skimage.exposure.equalize_adapthist])
# Plot animated gifj
plt.close()
Image(filename='animated_timeseries.gif')
Applying custom image processing functions
Exporting animation to animated_timeseries.gif
[17]:
<IPython.core.display.Image object>
Additional information¶
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Africa data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Slack channel or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
Github.
Compatible datacube version:
[18]:
print(datacube.__version__)
1.8.15
Last Tested:
[19]:
from datetime import datetime
datetime.today().strftime('%Y-%m-%d')
[19]:
'2023-08-11'